Cells contain DNA which provide the basic instructions for biological cell function and therefore, life as we know it. If the DNA within a single cell was taken out and stretched end to end, it would measure in the region of 2 metres (Piovesan et al 2019). Taken up to the multicellular organismal scale, this provides a massive target which is at risk of being damaged, resulting in DNA damage.
Multiple forms of DNA damage have been characterised (Chatterjee and Walker, 2017). These are induced and influenced by multiple sources, from carcinogens and drugs to environmental factors and faulty DNA repair mechanisms. This can lead to faulty DNA replication and erroneous cell function, which plays a role in the development of cancer (Phillips and Arlt, 2009), and has additionally been implicated in neural disorders. Faulty DNA repair mechanisms induce the expansion of trinucleotide repeats in the Huntingtin gene which directly correlates to age of onset of Huntingdon’s disease (Holmans et al., 2017). To counter DNA damage, the cell possesses a variety of repair mechanisms; several different pathways exist, comprised of multiple DNA repair enzymes (Pailas et al., 2022). Damage leads to DNA strand breakage (both single strand and double strand breaks). Quantification of DNA strand breakage and its subsequent repair allows the experimental scientist to assess the impact of a wide variety of factors and scenarios on DNA integrity.
Many modern molecular biology and sequencing techniques are now focused on analysing genomic data at the single cell level, which has revolutionised biology. To this author, who was an undergraduate 30 years ago, this is an almost space-age jump in progress. However, toxicologists have been performing single cell DNA analysis of DNA integrity for many years by means of the elegant COMET assay. The COMET assay, or single cell gel electrophoresis (SCGE), is one of the simplest and most sensitive methods of assessing DNA damage (Olive et al., 1990; Singh et al, 1988). It is relatively easy to perform and requires little specialised equipment, other than a gel tank and a fluorescence microscope (or other equipment capable of imaging fluorescent dyes).
In simple terms, the COMET assay involves embedding cells in thin layers of agarose coated on a microscope slide and then performing a cell lysis step. DNA within the cell nucleus is then allowed to unwind under alkaline or neutral conditions. It is subsequently separated by electrophoresis, prior to being stained and imaged using a UV microscope. If the DNA is damaged then it can migrate out of the cell in an electric current in the same manner that linearised DNA fragments of different molecular weights are separated on an agarose gel. Upon staining with fluorescent intercalating dyes such as ethidium bromide (or more ‘friendly’ modern alternatives such as GelRed), the damaged DNA can be visualised and it resembles a comet with the tail spreading away from the cell nucleus. Undamaged DNA remains within the cell nucleus and so exhibits no COMET tail. The distance that the DNA migrates from the cell is quantifiable and directly proportional to the amount of damage sustained (e.g., strand breakages).
Measurements of COMET damage are made using computer-based image analysis and include % tail DNA, tail moment (tail intensity multiplied by tail length), and olive moment (% of DNA within the tail multiplied by the length between the centres of the head and tail). Classically, % tail DNA and moment have been employed most frequently; however, olive moment is increasing in popularity as it is thought to be superior at describing the heterogeneity and variation in DNA distribution within the COMET tail throughout a population of cells (Kumaravel et al., 2009). Modification of the buffers involved allows different types of damage to be detected; in neutral COMET assays, the double stranded structure of DNA is maintained and the assay measures double stranded breaks, whereas in the alkaline COMET assay, double stranded breaks, single stranded breaks and alkaline-labile sites are all assessed and so the assay is more sensitive (Peycheva et al., 2014; Lu et al., 2017).
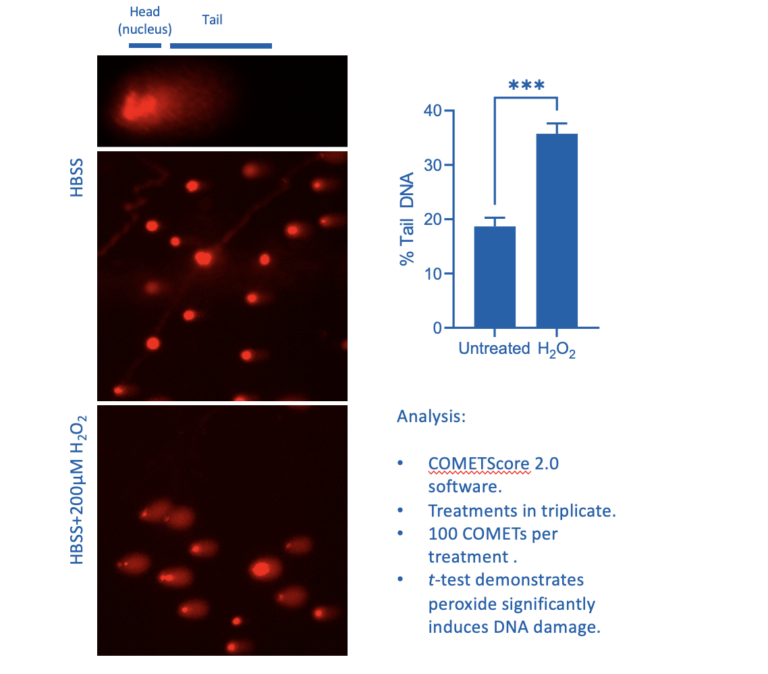
Figure 1: Influence of 20 minutes H2O2 treatment on HCT116 COMET formation
Figure 1 illustrates the difference in COMETs between untreated cells and cells where oxidative stress has resulted in DNA strand breakages following treatment with hydrogen peroxide for 20 minutes. The COMET-like appearance of the tail following the induction of DNA damage is quite striking. This basic assay may be expanded on to assess the ability of the cell to repair damage by using a pulse-chase model. Damage can be induced, a sample of cells taken to characterise 100% damage, and the damaging reagent can then be washed away to be replaced with normal culture media. Cells can be sampled at increasing times post-induction of damage in this pulse-chase experiment and COMET assays performed to assess levels of strand breakage. It can be surprising to the casual observer how quickly damage can be repaired, with significant repair being seen within 15 minutes (Singh et al., 1988).
Historically, the COMET assay was originally quite an intensive process. Frosted microscope slides had to be carefully layered with multiple layers of agarose prior to use and it was common for these to float off into the assay buffers. However, pre-made COMET slides are now available at a reasonable price from commercial suppliers such as R&D/Trevigen. Performing COMET assays is thus happier and less stressful as the cells embedded in LMP agarose adhere more successfully to these slides and are less prone to tearing when coverslips are removed. If you have ever had success in applying water-based transfers to AirFix models (or found them ripping when you tried to handle them) then you have experienced the deftness and delicacy of touch required to successfully work with these fine layers of agarose.
In addition to slide preparation, imaging and reading COMET data used to be time consuming, as each slide had to be read individually and images of COMETs collected manually on a fluorescence microscope. Over the years, the development of software has allowed multiple COMETs to be automatically scored on each microscope image, but in order to count 100+ COMETs per treatment, this was still an onerous task. Here at Cellomatics Biosciences, we have invested in high content screening equipment which has sped up the process of running COMET assays by enabling us to screen COMET slides en masse, combined with automated image analysis. So, this once exacting task is now simplified and a much speedier proposition.
As a footnote, the development of the COMET assay forges on ahead in other research arenas. Recent progress has given the researcher a potentially novel perspective by combining the COMET assay with FISH technology (fluorescent in situ hybridisation; Spivak et al., 2009). This has the potential to revolutionise COMET data by determining if specific DNA sequences are located in the nucleus or the damaged region of DNA forming the COMET tail.
At Cellomatics, our scientists are experts in a wide range of assay development and execution. Find out more here.
References
Chatterjee N and Walker GC (2017). Mechanisms of DNA damage, repair and mutagenesis. Environ. Mol. Mutagen. 58:235-263
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5474181/
Holmans PA, Massey TH, Jones L (2017). Genetic modifiers of Mendelian disease: Huntington’s disease and the trinucleotide repeat disorders. Hum. Mol. Genet. 26: R83-R90
https://pubmed.ncbi.nlm.nih.gov/28977442/
Kumaravel TS, Vilhar B, Faux SP, Jha AN (2009). COMET Assay measurements: A perspective. Cell Biol. Toxicol. 25: 53-64
https://link.springer.com/article/10.1007/s10565-007-9043-9
Lu Y, Liu Y and Yang, C (2017). Evaluating in vitro DNA damage using COMET assay. J. Vis. Exp. 128: e56450
https://www.jove.com/t/56450/evaluating-in-vitro-dna-damage-using-comet-assay
Olive PL, Banath JP and Durand RE (1990). Heterogeneity in radiation-induced DNA damage and repair in tumour and normal cells measured using the “comet” assay. Radiat. Res. 122: 86-94
https://pubmed.ncbi.nlm.nih.gov/2320728/
Pailas A, Niaka K, Zorzompokou C, Marangos P (2022). The DNA damage response in fully grown mammalian Oocytes. Cells 11: 798-814
https://www.mdpi.com/2073-4409/11/5/798
Peycheva E, Georgieva M, Miloshev G (2009). Comparison between alkaline and neutral variants of yeast COMET assay. Biotechnology & Biotechnological Equipment 23: 1090-1092
https://www.tandfonline.com/doi/abs/10.1080/13102818.2009.10817618
Phillips DH and Arlt VM (2009). Genotoxicity: damage to DNA and its consequences. EXS 99: 87-110
https://pubmed.ncbi.nlm.nih.gov/19157059/
Piovesan A, Pelleri MC, Antonaros F, Strippoli P, Caracausi M, Vitale L (2019). On the length, weight and GC content of the human genome. BMC Research Notes 12:106
https://bmcresnotes.biomedcentral.com/articles/10.1186/s13104-019-4137-z
Singh NP, McCoy MT, Tice RR and Schneider, EL (1988). A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175: 184-191
https://www.sciencedirect.com/science/article/abs/pii/0014482788902650
Spivak G, Cox RA and Hanawalt PC (2008). New applications of the COMET assay: COMET-FISH and transcription-coupled DNA repair. Mut. Res. 681: 44-50